This article and associated images are based on a poster originally authored by Rebecca L. Rich, Anthony M. Giannetti, John Rosenfeld, Adam Shutes and Yusuke Kawase and presented at ELRIG Drug Discovery 2025 in affiliation with Carterra, CarnaBio USA, Inc. and Carna Biosciences, Inc.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Introduction
As a drug target class, kinases continue to provide a wealth of opportunities for addressing human disease, but often can be challenging to work with in vitro. Additionally, the ubiquitous nature of kinases across many critical pathways means therapeutic targeting of this class necessitates careful consideration regarding off-target profiles. Direct label-free approaches, such as SPR, can complement activity assays by providing the intrinsic affinity and real-time kinetics of interactions. Here we highlight the power of combining an extensive panel of ready-made biotinylated kinases with HT-SPR to generate a wealth of compound binding information. In three days over 125,000 interactions were measured between a panel of kinases and the Maybridge 1000 fragment library. We also profiled a kinase-focused small molecule library and obtained more than 80,000 binding interactions in a three-day instrument run. Detailed kinetics were then subsequently obtained for hits of interest. Beyond simple yes/no reporting, this approach allows for nuanced kinetic profiling for up to hundreds of binding events in parallel, thereby enabling thoughtful discovery of safe and efficacious drug candidates.
Methods and materials
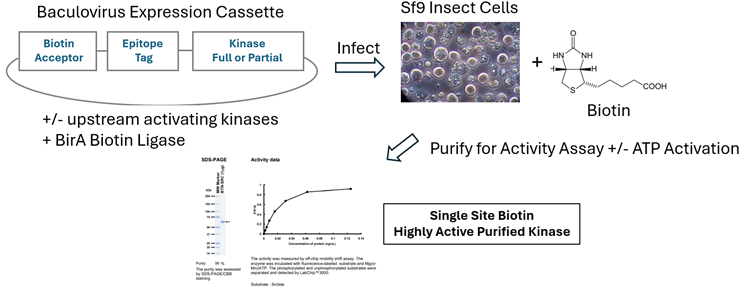
Figure 1. Production Schema for Highly Validated Single Site Biotinylated Active Kinases. Target kinases are cloned and expressed by baculovirus infection of Sf9 cells. Activated, single-site biotinylated kinases are purified using an epitope tag. Purity and degree of biotinylation are measured followed by activity assessment using a mobility shift assay or fluorescence polarization. Active forms of kinases are obtained by incubation with ATP and/or upstream activating kinases. CarnaBio produces over 200 BTN-Kinases. Image Credit: Image courtesy of Rebecca L. Rich et al., in partnership with ELRIG (UK) Ltd.
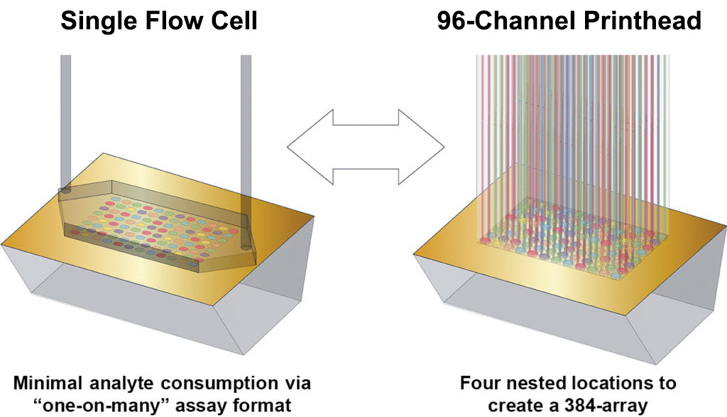
Figure 2. The Carterra line of instruments utilize a 96-channel print head to immobilize an array of ligands in up to 192- (Ultra) or 384-(XT) positions. The entire array is then docked into a single-flow cell system and analytes are injected sequentially yielding data on up to hundreds of ligand and reference spots simultaneously. Image Credit: Image courtesy of Rebecca L. Rich et al., in partnership with ELRIG (UK) Ltd.
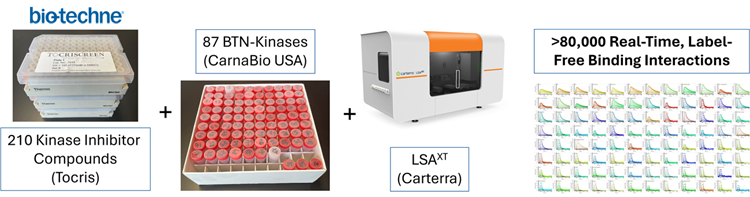
Figure 3. Kinetics Workflow for Inhibitor Compounds Binding to a Kinase Array. Binding studies were performed at 15 °C using the Carterra LSAXT HT-SPR biosensor. Multiple densities of each kinase and off-target proteins (in HBS, 0.005 % Tween-20, 5 % glycerol, 0.5 mg/mL BSA, pH 7.4) were captured at 384 locations on an SAD200M sensor chip. The Tocriscreen™ Kinase Inhibitor 3.0 library (Cat. No. 7844) was screened at 1 μM (in HBS, 0.005 % Tween-20, 5 % glycerol, 5 mM MgCl2,1 mm DTT, 3 % DMSO, pH 7.4) for binding to the kinase panel. Selected inhibitors were re-tested in a two-fold dilution series starting at concentrations up to 2 μM. Image Credit: Image courtesy of Rebecca L. Rich et al., in partnership with ELRIG (UK) Ltd.
Results

Figure 4. ~250 SPR Binding Cycle Sensorgrams. Each column depicts the responses for one compound binding to the array of 90 biotinylated kinases and controls, with individually colored sensorgrams representing a unique kinase. Image Credit: Image courtesy of Rebecca L. Rich et al., in partnership with ELRIG (UK) Ltd.
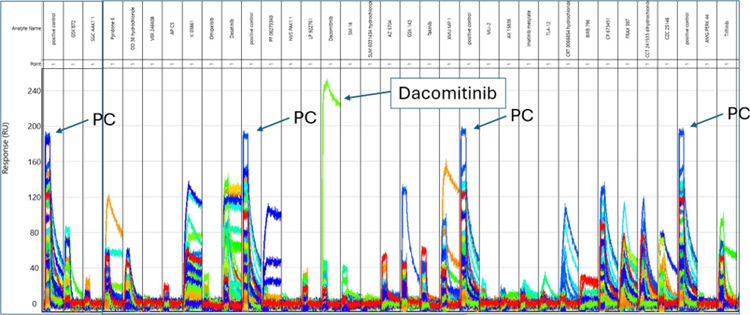
Figure 5. Zoomed-In View of Select Binding Cycles. Positive control (PC) compound CP673451 was injected every twelve cycles to monitor activity across the array of biotinylated kinases. Dacomitinib is highlighted as an example of a compound shown to specifically bind with high affinity to a single kinase. Image Credit: Image courtesy of Rebecca L. Rich et al., in partnership with ELRIG (UK) Ltd.
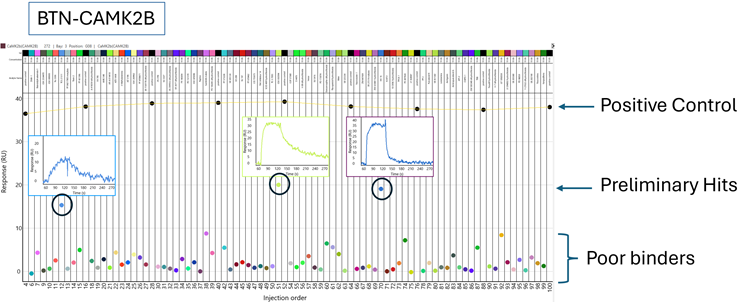
Figure 6. Binding Responses for Compounds against CAMK2B. Using 1-min association and 5-min dissociation phases, the positive control compound, CP673451, was tested periodically at 10 μM and the compound library was screened at 1 μM. Repetition of the control compound shows consistency and durability of CAMK2B activity over time. Several clear hits are indicated. Image Credit: Image courtesy of Rebecca L. Rich et al., in partnership with ELRIG (UK) Ltd.
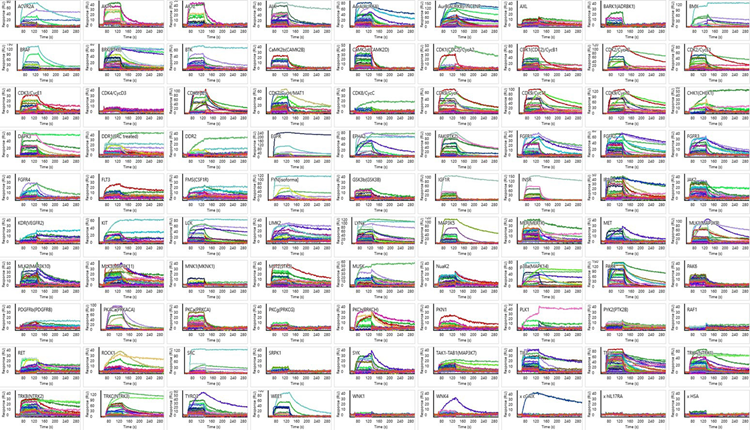
Figure 7. Subset of 80 Compounds Binding to 87 Kinases. Each tile plot corresponds to a single kinase, with each colored curve representing a distinct compound binding profile. A diverse range of weak to very stable interactions are demonstrated. Three negative–control proteins are included in the bottom right. Image Credit: Image courtesy of Rebecca L. Rich et al., in partnership with ELRIG (UK) Ltd.
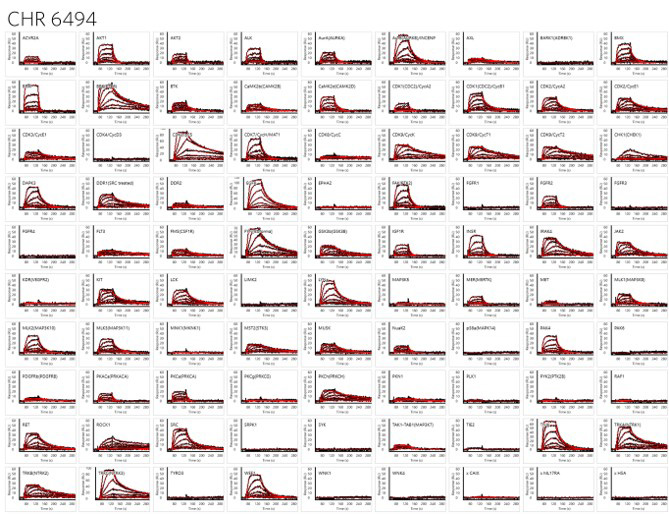
Figure 8. Detailed Kinetic Fingerprints of CHR 6494 (415 Da). A single set of dose-response injections provides detailed affinity and kinetic data for 90 kinases simultaneously. Image Credit: Image courtesy of Rebecca L. Rich et al., in partnership with ELRIG (UK) Ltd.
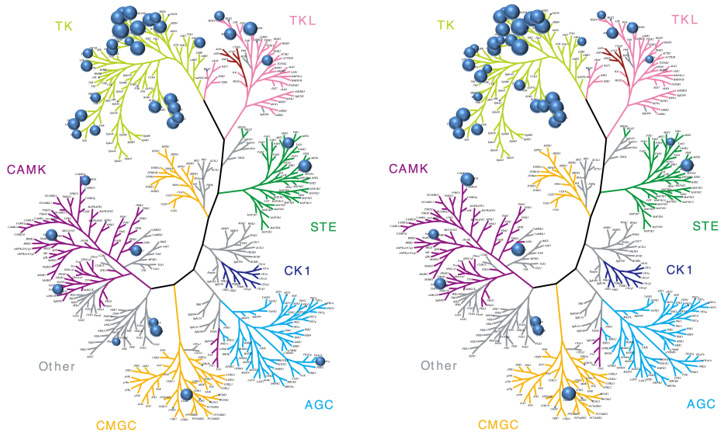
Figure 9. Kinome profiling of sunitinib (Sutent). Left: SPR-derived affinities (expressed as pKD). Right: pIC50s at 1 μM from Carna’s mobility shift assay. Results are shown on kinome phylogenetic trees where larger bubbles represent more potent “hits”. Sunitinib is a multi-targeted tyrosine kinase inhibitor, and used in the clinic to inhibit PDGFR, FGFRs, VEGFR, and PDGFR. There is a strong concordance between the SPR KDs and the EZ-Reader off-chip MSA assays across families of kinases. Image Credit: Image courtesy of Rebecca L. Rich et al., in partnership with ELRIG (UK) Ltd.
High throughput fragment screening using Carterra Ultra

Figure 10. The new Carterra Ultra™ can print one reference spot next to each ligand spot for maximum sensitivity. Image Credit: Image courtesy of Rebecca L. Rich et al., in partnership with ELRIG (UK) Ltd.
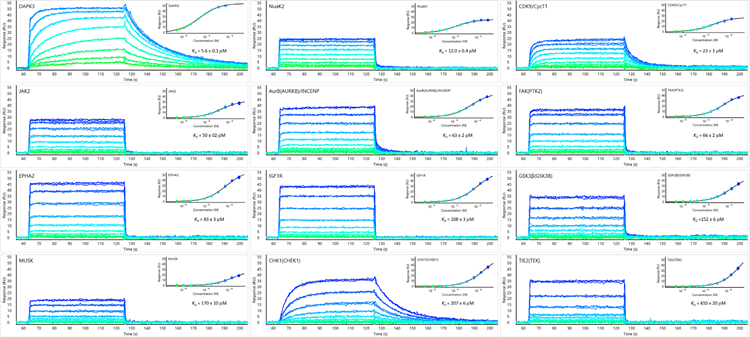
Figure 11. Data From Carterra Ultra: Triplicate overlays of AMP-PNP binding to 12 kinases with affinities ranging from 5-400 μM. Kinetics for even very weak associations is readily observed. Image Credit: Image courtesy of Rebecca L. Rich et al., in partnership with ELRIG (UK) Ltd.
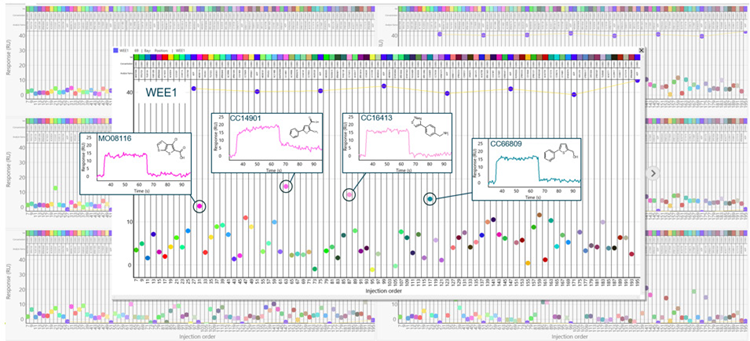
Figure 12. Binding report points vs. cycle number for seven kinases. Hits with binding levels exceeding the background scatter for WEE1 are highlighted. Image Credit: Image courtesy of Rebecca L. Rich et al., in partnership with ELRIG (UK) Ltd.

Figure 13. Fragment screening against 96 proteins. One compound binds only two proteins (top panel) while one compound binds only the carbonic anhydrase control spots (bottom panel). Image Credit: Image courtesy of Rebecca L. Rich et al., in partnership with ELRIG (UK) Ltd.
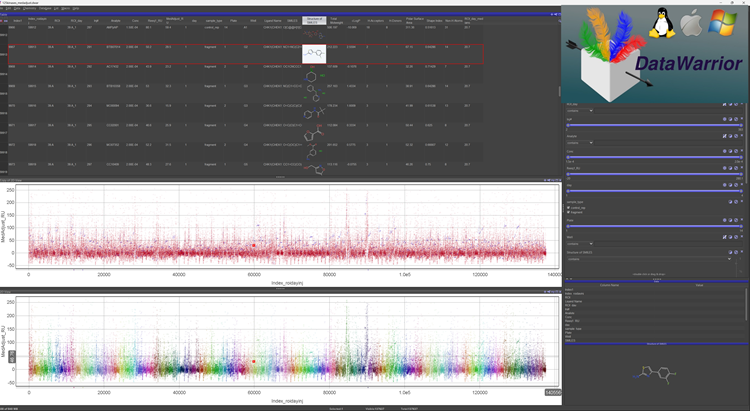
Figure 14. Fragment screen of the Maybridge 1000 library against a panel of 125 BTN-kinases. ~150,000 interactions with fragments and controls were measured on a single instrument in three days. The screen is visualized in DataWarrior (openmolecules.org). The middle panel shows fragments (red) and controls (blue). Each color in the lower panel represents one complete 1,000 compound screen against a single kinase (125 in all). The red dot indicates CHK1 binding to the shown difluoroaminothiazole compound. Image Credit: Image courtesy of Rebecca L. Rich et al., in partnership with ELRIG (UK) Ltd.
Conclusions
- Carterra’s HT-SPR solutions deliver data on >20-50 fold more ligands than other SPR-platforms
- Ultra’s noise, stability, and thermal control deliver quality binding data in support of small molecule hit-to-lead including fragment screening
- Using CarnaBio’s single-site biotinylated active kinases enables easy assay development with plug-and-play potential for any combination of up to hundreds of kinases and off-targets using HT-SPR.
About Carterra, Inc.
Carterra® is a leading provider of innovative technologies designed to accelerate the discovery of novel therapeutic candidates. Carterra’s high throughput LSA® instrument for monoclonal antibody (mAb) screening and characterization combines patented microfluidics technology with real-time High Throughput Surface Plasmon Resonance (HT-SPR) and industry leading Kinetic and Epitope analysis and visualization software, delivering up to 100 times the throughput in 10% of the time while using only 1% of the sample compared to existing platforms.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free to attend!
Our values
Our values are to always ensure the highest quality of content, that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer-led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world-class conferences, networking events, webinars, and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Jan 6, 2026